We are glad to present an open ecosystem to foster the exchange of machine learning modules in the scientific community: the DEEP Marketplace. The catalogue includes all the applications developed in the DEEP Hybrid DataCloud project (DEEP), as well as external modules developed by users who want to share their applications and solution approaches within the AI community, thus promoting collaboration between research and development (R&D) groups.
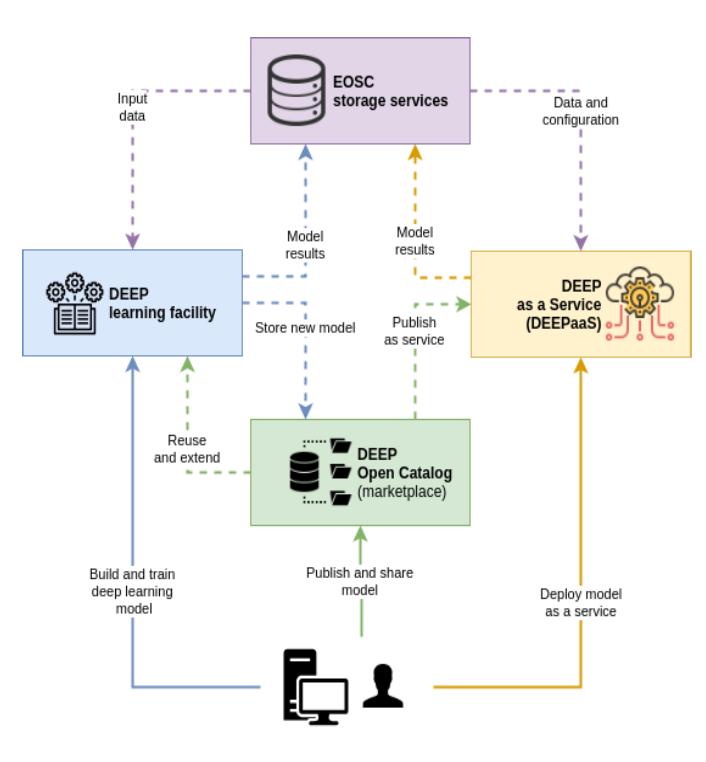
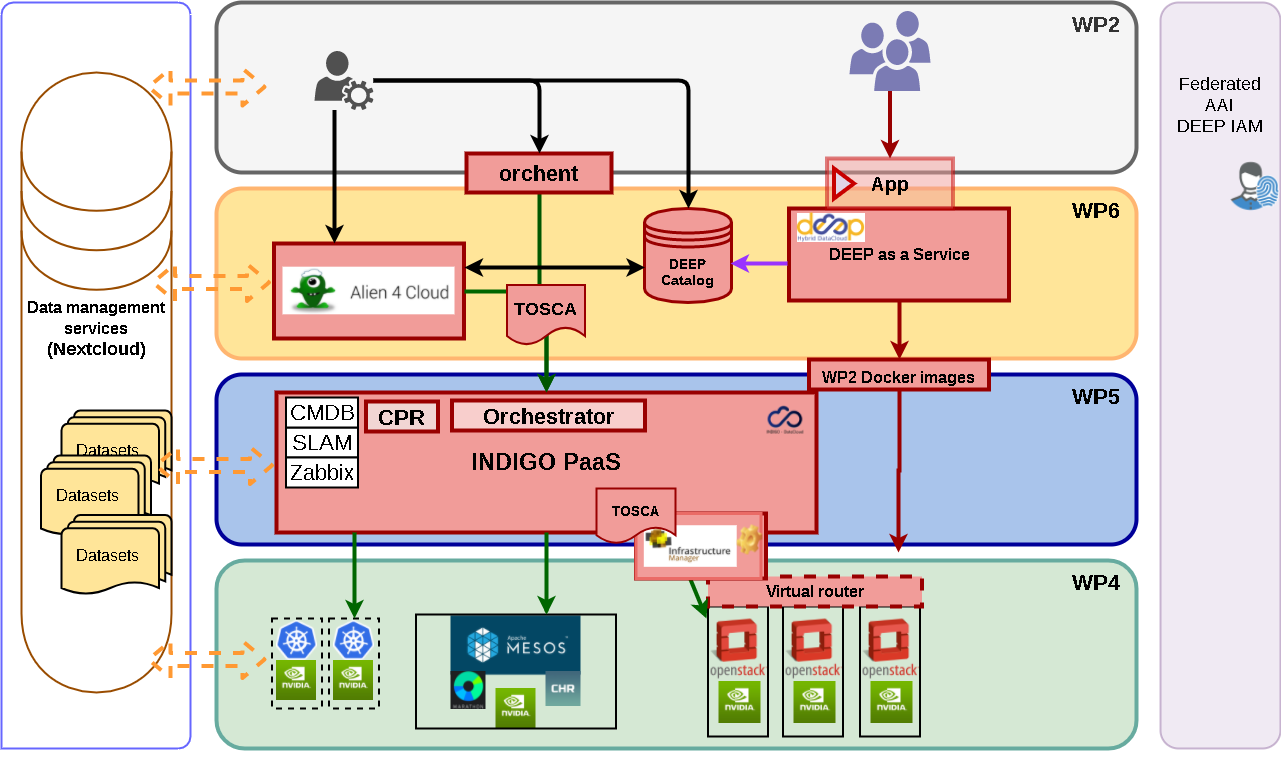
The DEEP framework supports artificial intelligent modules, especially compute-intensive deep learning modules over distributed e-infrastructures in the European Open Science Cloud (EOSC) for the most compute-intensive tasks of the intelligent software development and deployment life-cycle phases. Moreover, the DEEP Open Catalogue (available in DEEP Marketplace), DEEP as Service solution and DEEP learning facilities are in connection with the EOSC storage and data services, which allow to easily share, publish, exchange, collaborate and deploy developed models as services. These, in turn, support Open Science in which transparency and reproducibility of computational work are important.
DEEP use cases as deep learning modules are publicly available as Open Source in the DEEP Open Catalogue and publicly accessible in the DEEP Open Market. They are categorized in the following main extendable groups:
- Earth observations: deep neural network applications to perform pattern recognition on satellite images. They can be combined with other in-situ measurements for ecosystems and biodiversity to perform tasks such as remote object detection, terrain segmentation or meteorological prediction. Currently we offer a super-resolution module to upscale low resolution bands to high resolution for the most popular multispectral satellites from all around the world.
- Biological and medical science: deep learning modules for biomedical image analysis have opened new opportunities in how diseases are diagnosed and treated. We currently provide a retinopathy automated classification based on color fundus retinal photography images.
- Cyber-security and network monitoring: we provide modules with co-functions enhancements for surveillance Intrusion Detection Systems supervising traffic network flows of computing infrastructure.
- Citizen science: deep learning modules for leveraging citizen science in large-scale biodiversity monitoring. The available modules include automatic identification of species from images for a wide range of categories (plants, seeds, conus and phytoplankton). Most of these modules are also available as mobile applications.
- General purpose: this includes modules that can be used across a wide range of domains. We currently provide modules for image classification, audio classification, speech-to-text synthesis and pose detection.
These groups are open extensible categories, which offer flexible way for adding new and interesting applications to be presented in the DEEP Marketplace for wide and public audience.
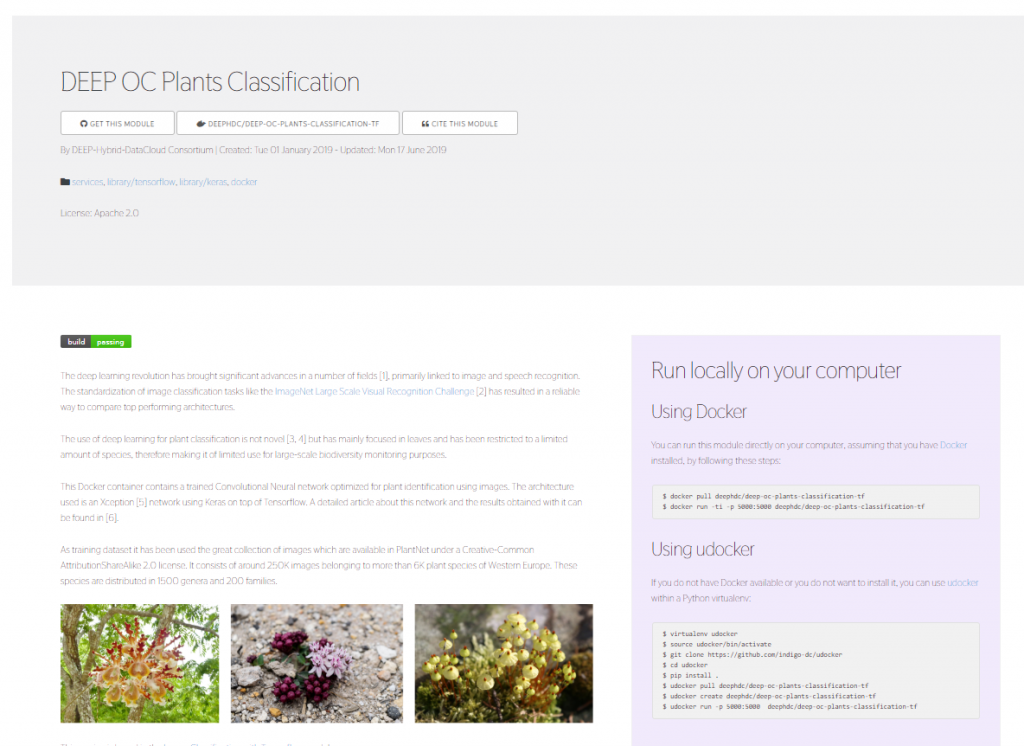
Figure 1. A snapshot of the application catalogue.
All modules in DEEP Open Catalogue is categorized by name and by tags like #tensorflow, #keras or #docker with upcoming full-text search capability. For every module a set of metadata entries are provided. The full list of metadata entries includes:
- Title
- Summary
A quick one liner describing the module
- Description
An extended description of what the module performs, what problem is it solving, what machine learning tools it is using, which data types it needs for prediction, etc.
- Tags
These are the tags that are going to make the module more searchable. Tags usually refer to overall features or tools used in the module.
- License
Under what license a module can be used, modified, and shared. - Creation date
- “Cite this module” – URL for citation (for modules based on a published paper)
URL pointing to the DOI of the paper
- “Get this module” – URL for the module’s source code
This points to the source code that is used to run the application. If you plan to use the application (especially if you want to retrain it) it always good to have a look to the README in this repository as it is going to explain in detail what you need to successfully retrain the application.
- URL for the module’s Docker image in Dockerhub
This is the Docker image of the module. It can be hosted in the deephdc organization or somewhere else.
- URL for training weights of the module
This is usually a compressed file with all the info from the training (including the final weights). Together with the module’s code, it should be enough to reproduce any of the model’s functionality.
- URL pointing to the dataset used for training
This points to the dataset used for training (for example ImageNet for some image classification modules)
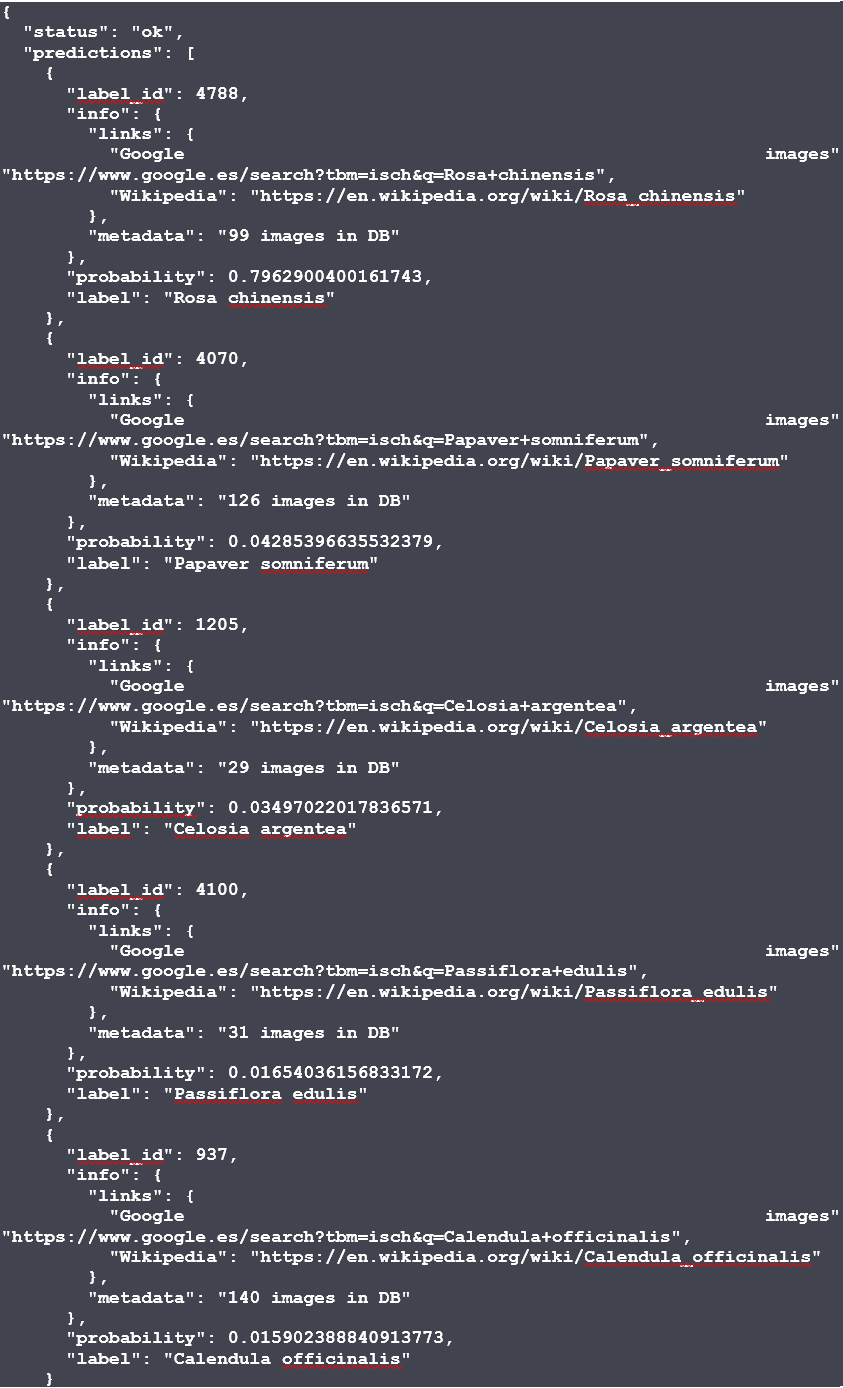
Not all the modules need to define all the metadata fields. In the module’s page you will also see a small side panel with quick one-liner instructions on how to run the module with Docker and start making predictions: a simple example would be to run the Docker container locally and go to http://127.0.0.1:5000/docs in your local browser to interact with the API.
As you can see, we provide all the tools to make the code exportable to any platform as the code is completely open source and not linked to the DEEP infrastructure in any way. Any user can access the code of any application to modify it freely.
As explained in the documentation modules in the Marketplace modules are intended for two purposes:
- So that a user uses a module “as it is” for prediction.
- So that the user finetunes the module to a specific task. For example users reuse the image classification module to train a classifier for x-ray scans.
Sometimes users will find that their task requires code that is not present in any of the available modules’ functionalities. So users can develop their own custom code from scratch using the DEEP template if they want to, or starting from another module’s code as all modules have Open Source Licenses. Once users have their new module developed to perform the desired task they can request to upload it to the Marketplace to share it with the community. For detailed up-to-date instructions on all these steps please visit the DEEP documentation.
So, with all of this, we have covered the basics of the Marketplace:
- How to browse for modules
- How to browse the metadata inside the module
- How modules are structured: module’s code, container’s code, weights, etc.
- How a user can deploy the modules either for predict or to retrain
- How user’s can develop their own modules to perform a task that is not available in the Marketplace. These new modules can then be shared with the whole community.